31 KiB
| title | teaser | menu | next | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Layers and Model Architectures | Power spaCy components with custom neural networks |
|
/usage/projects |
Example
from thinc.api import Model, chain @spacy.registry.architectures.register("model.v1") def build_model(width: int, classes: int) -> Model: tok2vec = build_tok2vec(width) output_layer = build_output_layer(width, classes) model = chain(tok2vec, output_layer) return model
A model architecture is a function that wires up a
Thinc Model instance. It describes the
neural network that is run internally as part of a component in a spaCy
pipeline. To define the actual architecture, you can implement your logic in
Thinc directly, or you can use Thinc as a thin wrapper around frameworks such as
PyTorch, TensorFlow and MXNet. Each Model can also be used as a sublayer of a
larger network, allowing you to freely combine implementations from different
frameworks into a single model.
spaCy's built-in components require a Model instance to be passed to them via
the config system. To change the model architecture of an existing component,
you just need to update the config so that it refers
to a different registered function. Once the component has been created from
this config, you won't be able to change it anymore. The architecture is like a
recipe for the network, and you can't change the recipe once the dish has
already been prepared. You have to make a new one.
### config.cfg (excerpt)
[components.tagger]
factory = "tagger"
[components.tagger.model]
@architectures = "model.v1"
width = 512
classes = 16
Type signatures
Example
from typing import List from thinc.api import Model, chain from thinc.types import Floats2d def chain_model( tok2vec: Model[List[Doc], List[Floats2d]], layer1: Model[List[Floats2d], Floats2d], layer2: Model[Floats2d, Floats2d] ) -> Model[List[Doc], Floats2d]: model = chain(tok2vec, layer1, layer2) return model
The Thinc Model class is a generic type that can specify its input and
output types. Python uses a square-bracket notation for this, so the type
Model[List, Dict] says that each batch of inputs to the model will be a
list, and the outputs will be a dictionary. You can be even more specific and
write for instanceModel[List[Doc], Dict[str, float]] to specify that the
model expects a list of Doc objects as input, and returns a
dictionary mapping of strings to floats. Some of the most common types you'll
see are:
| Type | Description |
|---|---|
A batch of Doc objects. Most components expect their models to take this as input. |
|
A two-dimensional numpy or cupy array of floats. Usually 32-bit. |
|
A two-dimensional numpy or cupy array of integers. Common dtypes include uint64, int32 and int8. |
|
A list of two-dimensional arrays, generally with one array per Doc and one row per token. |
|
| A container to handle variable-length sequence data in an unpadded contiguous array. | |
| A container to handle variable-length sequence data in a padded contiguous array. |
See the Thinc type reference for details. The
model type signatures help you figure out which model architectures and
components can fit together. For instance, the
TextCategorizer class expects a model typed
Model[List[Doc], Floats2d], because the model will predict one row of
category probabilities per Doc. In contrast, the
Tagger class expects a model typed Model[List[Doc],
List[Floats2d]], because it needs to predict one row of probabilities per
token.
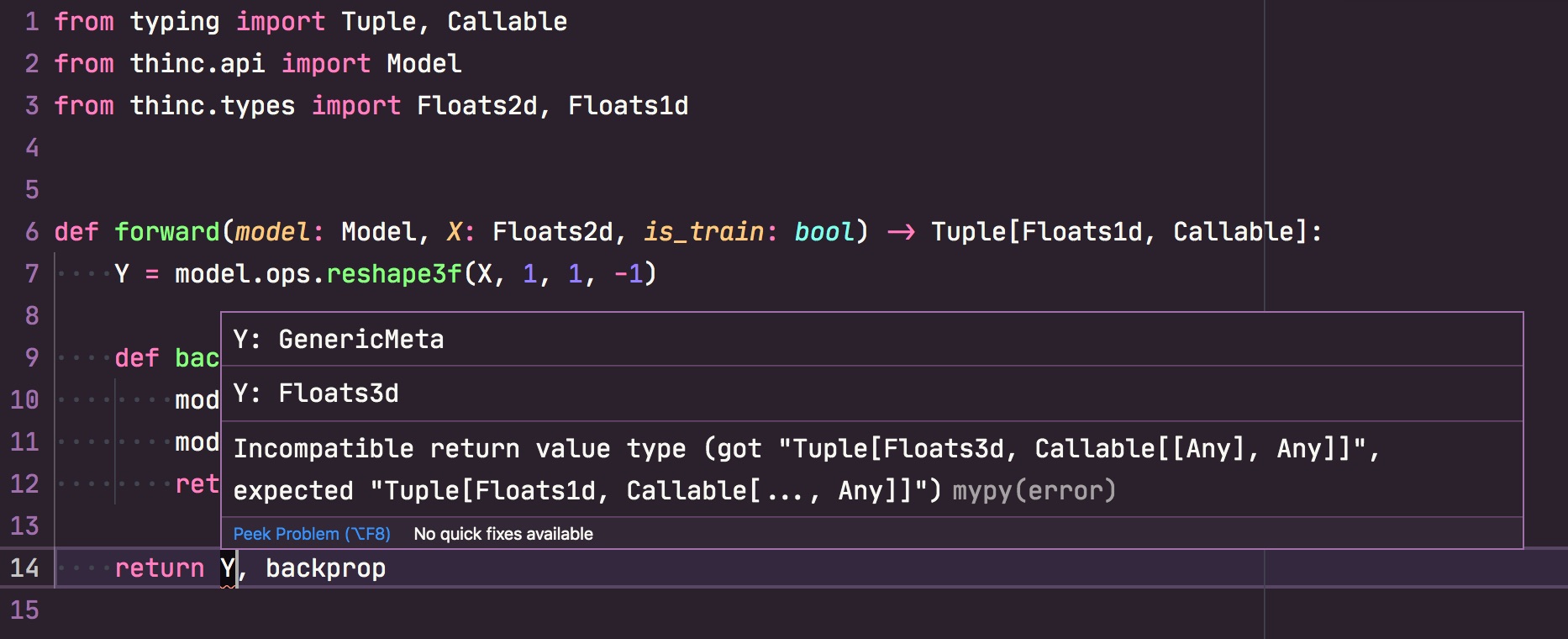
There's no guarantee that two models with the same type signature can be used interchangeably. There are many other ways they could be incompatible. However, if the types don't match, they almost surely won't be compatible. This little bit of validation goes a long way, especially if you configure your editor or other tools to highlight these errors early. The config file is also validated at the beginning of training, to verify that all the types match correctly.
If you're using a modern editor like Visual Studio Code, you can
set up mypy with the
custom Thinc plugin and get live feedback about mismatched types as you write
code.
Swapping model architectures
If no model is specified for the TextCategorizer, the
TextCatEnsemble architecture is used by
default. This architecture combines a simple bag-of-words model with a neural
network, usually resulting in the most accurate results, but at the cost of
speed. The config file for this model would look something like this:
### config.cfg (excerpt)
[components.textcat]
factory = "textcat"
labels = []
[components.textcat.model]
@architectures = "spacy.TextCatEnsemble.v1"
exclusive_classes = false
pretrained_vectors = null
width = 64
conv_depth = 2
embed_size = 2000
window_size = 1
ngram_size = 1
dropout = 0
nO = null
spaCy has two additional built-in textcat architectures, and you can easily
use those by swapping out the definition of the textcat's model. For instance,
to use the simple and fast bag-of-words model
TextCatBOW, you can change the config to:
### config.cfg (excerpt) {highlight="6-10"}
[components.textcat]
factory = "textcat"
labels = []
[components.textcat.model]
@architectures = "spacy.TextCatBOW.v1"
exclusive_classes = false
ngram_size = 1
no_output_layer = false
nO = null
For details on all pre-defined architectures shipped with spaCy and how to configure them, check out the model architectures documentation.
Defining sublayers
Model architecture functions often accept sublayers as arguments, so that you can try substituting a different layer into the network. Depending on how the architecture function is structured, you might be able to define your network structure entirely through the config system, using layers that have already been defined.
In most neural network models for NLP, the most important parts of the network
are what we refer to as the
embed and encode steps.
These steps together compute dense, context-sensitive representations of the
tokens, and their combination forms a typical
Tok2Vec layer:
### config.cfg (excerpt)
[components.tok2vec]
factory = "tok2vec"
[components.tok2vec.model]
@architectures = "spacy.Tok2Vec.v1"
[components.tok2vec.model.embed]
@architectures = "spacy.MultiHashEmbed.v1"
# ...
[components.tok2vec.model.encode]
@architectures = "spacy.MaxoutWindowEncoder.v1"
# ...
By defining these sublayers specifically, it becomes straightforward to swap out a sublayer for another one, for instance changing the first sublayer to a character embedding with the CharacterEmbed architecture:
### config.cfg (excerpt)
[components.tok2vec.model.embed]
@architectures = "spacy.CharacterEmbed.v1"
# ...
[components.tok2vec.model.encode]
@architectures = "spacy.MaxoutWindowEncoder.v1"
# ...
Most of spaCy's default architectures accept a tok2vec layer as a sublayer
within the larger task-specific neural network. This makes it easy to switch
between transformer, CNN, BiLSTM or other feature extraction approaches. The
transformers documentation
section shows an example of swapping out a model's standard tok2vec layer with
a transformer. And if you want to define your own solution, all you need to do
is register a Model[List[Doc], List[Floats2d]] architecture function, and
you'll be able to try it out in any of the spaCy components.
Wrapping PyTorch, TensorFlow and other frameworks
Thinc allows you to wrap models
written in other machine learning frameworks like PyTorch, TensorFlow and MXNet
using a unified Model API. This makes it
easy to use a model implemented in a different framework to power a component in
your spaCy pipeline. For example, to wrap a PyTorch model as a Thinc Model,
you can use Thinc's
PyTorchWrapper:
from thinc.api import PyTorchWrapper
wrapped_pt_model = PyTorchWrapper(torch_model)
Let's use PyTorch to define a very simple neural network consisting of two
hidden Linear layers with ReLU activation and dropout, and a
softmax-activated output layer:
### PyTorch model
from torch import nn
torch_model = nn.Sequential(
nn.Linear(width, hidden_width),
nn.ReLU(),
nn.Dropout2d(dropout),
nn.Linear(hidden_width, nO),
nn.ReLU(),
nn.Dropout2d(dropout),
nn.Softmax(dim=1)
)
The resulting wrapped Model can be used as a custom architecture as such,
or can be a subcomponent of a larger model. For instance, we can use Thinc's
chain combinator, which works like
Sequential in PyTorch, to combine the wrapped model with other components in a
larger network. This effectively means that you can easily wrap different
components from different frameworks, and "glue" them together with Thinc:
from thinc.api import chain, with_array, PyTorchWrapper
from spacy.ml import CharacterEmbed
wrapped_pt_model = PyTorchWrapper(torch_model)
char_embed = CharacterEmbed(width, embed_size, nM, nC)
model = chain(char_embed, with_array(wrapped_pt_model))
In the above example, we have combined our custom PyTorch model with a character
embedding layer defined by spaCy.
CharacterEmbed returns a Model that takes
a List[Doc] as input, and outputs a List[Floats2d]. To make sure that
the wrapped PyTorch model receives valid inputs, we use Thinc's
with_array helper.
You could also implement a model that only uses PyTorch for the transformer layers, and "native" Thinc layers to do fiddly input and output transformations and add on task-specific "heads", as efficiency is less of a consideration for those parts of the network.
Using wrapped models
To use our custom model including the PyTorch subnetwork, all we need to do is
register the architecture using the
architectures registry. This assigns the
architecture a name so spaCy knows how to find it, and allows passing in
arguments like hyperparameters via the config. The
full example then becomes:
### Registering the architecture {highlight="9"}
from typing import List
from thinc.types import Floats2d
from thinc.api import Model, PyTorchWrapper, chain, with_array
import spacy
from spacy.tokens.doc import Doc
from spacy.ml import CharacterEmbed
from torch import nn
@spacy.registry.architectures("CustomTorchModel.v1")
def create_torch_model(
nO: int,
width: int,
hidden_width: int,
embed_size: int,
nM: int,
nC: int,
dropout: float,
) -> Model[List[Doc], List[Floats2d]]:
char_embed = CharacterEmbed(width, embed_size, nM, nC)
torch_model = nn.Sequential(
nn.Linear(width, hidden_width),
nn.ReLU(),
nn.Dropout2d(dropout),
nn.Linear(hidden_width, nO),
nn.ReLU(),
nn.Dropout2d(dropout),
nn.Softmax(dim=1)
)
wrapped_pt_model = PyTorchWrapper(torch_model)
model = chain(char_embed, with_array(wrapped_pt_model))
return model
The model definition can now be used in any existing trainable spaCy component, by specifying it in the config file. In this configuration, all required parameters for the various subcomponents of the custom architecture are passed in as settings via the config.
### config.cfg (excerpt) {highlight="5-5"}
[components.tagger]
factory = "tagger"
[components.tagger.model]
@architectures = "CustomTorchModel.v1"
nO = 50
width = 96
hidden_width = 48
embed_size = 2000
nM = 64
nC = 8
dropout = 0.2
Remember that it is best not to rely on any (hidden) default values, to ensure that training configs are complete and experiments fully reproducible.
Note that when using a PyTorch or Tensorflow model, it is recommended to set the
GPU memory allocator accordingly. When gpu_allocator is set to "pytorch" or
"tensorflow" in the training config, cupy will allocate memory via those
respective libraries, preventing OOM errors when there's available memory
sitting in the other library's pool.
### config.cfg (excerpt)
[training]
gpu_allocator = "pytorch"
Custom models with Thinc
Of course it's also possible to define the Model from the previous section
entirely in Thinc. The Thinc documentation provides details on the
various layers and helper functions
available. Combinators can be used to
overload operators and a common
usage pattern is to bind chain to >>. The "native" Thinc version of our
simple neural network would then become:
from thinc.api import chain, with_array, Model, Relu, Dropout, Softmax
from spacy.ml import CharacterEmbed
char_embed = CharacterEmbed(width, embed_size, nM, nC)
with Model.define_operators({">>": chain}):
layers = (
Relu(hidden_width, width)
>> Dropout(dropout)
>> Relu(hidden_width, hidden_width)
>> Dropout(dropout)
>> Softmax(nO, hidden_width)
)
model = char_embed >> with_array(layers)
Note that Thinc layers define the output dimension (nO) as the first argument,
followed (optionally) by the input dimension (nI). This is in contrast to how
the PyTorch layers are defined, where in_features precedes out_features.
Shape inference in Thinc
It is not strictly necessary to define all the input and output dimensions
for each layer, as Thinc can perform
shape inference between
sequential layers by matching up the output dimensionality of one layer to the
input dimensionality of the next. This means that we can simplify the layers
definition:
Diff
layers = ( Relu(hidden_width, width) >> Dropout(dropout) - >> Relu(hidden_width, hidden_width) + >> Relu(hidden_width) >> Dropout(dropout) - >> Softmax(nO, hidden_width) + >> Softmax(nO) )
with Model.define_operators({">>": chain}):
layers = (
Relu(hidden_width, width)
>> Dropout(dropout)
>> Relu(hidden_width)
>> Dropout(dropout)
>> Softmax(nO)
)
Thinc can even go one step further and deduce the correct input dimension of
the first layer, and output dimension of the last. To enable this functionality,
you have to call
Model.initialize with an input
sample X and an output sample Y with the correct dimensions:
### Shape inference with initialization {highlight="3,7,10"}
with Model.define_operators({">>": chain}):
layers = (
Relu(hidden_width)
>> Dropout(dropout)
>> Relu(hidden_width)
>> Dropout(dropout)
>> Softmax()
)
model = char_embed >> with_array(layers)
model.initialize(X=input_sample, Y=output_sample)
The built-in pipeline components in spaCy ensure
that their internal models are always initialized with appropriate sample
data. In this case, X is typically a List[Doc], while Y is typically a
List[Array1d] or List[Array2d], depending on the specific task. This
functionality is triggered when nlp.initialize is
called.
Dropout and normalization in Thinc
Many of the available Thinc layers allow you
to define a dropout argument that will result in "chaining" an additional
Dropout layer. Optionally, you can
often specify whether or not you want to add layer normalization, which would
result in an additional
LayerNorm layer. That means that
the following layers definition is equivalent to the previous:
with Model.define_operators({">>": chain}):
layers = (
Relu(hidden_width, dropout=dropout, normalize=False)
>> Relu(hidden_width, dropout=dropout, normalize=False)
>> Softmax()
)
model = char_embed >> with_array(layers)
model.initialize(X=input_sample, Y=output_sample)
Create new trainable components
In addition to swapping out default models in built-in
components, you can also implement an entirely new,
trainable pipeline component
from scratch. This can be done by creating a new class inheriting from
Pipe, and linking it up to your custom model implementation.
For details on how to implement pipeline components, check out the usage guide
on custom components and the
overview of the Pipe methods used by
trainable components.
Example: Entity elation extraction component
This section outlines an example use-case of implementing a novel relation extraction component from scratch. We'll implement a binary relation extraction method that determines whether or not two entities in a document are related, and if so, what type of relation. We'll allow multiple types of relations between two such entities (multi-label setting). There are two major steps required:
- Implement a machine learning model specific to this
task. It will have to extract candidates from a
Docand predict a relation for the available candidate pairs. - Implement a custom pipeline component powered by the
machine learning model that sets annotations on the
Docpassing through the pipeline.
Step 1: Implementing the Model
We need to implement a Model that takes a
list of documents (List[Doc]) as input, and outputs a two-dimensional
matrix (Floats2d) of predictions:
Model type annotations
The
Modelclass is a generic type that can specify its input and output types, e.g.Model[List[Doc], Floats2d]. Type hints are used for static type checks and validation. See the section on type signatures for details.
### Register the model architecture
@registry.architectures.register("rel_model.v1")
def create_relation_model(...) -> Model[List[Doc], Floats2d]:
model = ... # 👈 model will go here
return model
The first layer in this model will typically be an
embedding layer such as a
Tok2Vec component or a Transformer. This
layer is assumed to be of type Model[List[Doc], List[Floats2d]] as it
transforms each document into a list of tokens, with each token being
represented by its embedding in the vector space.
Next, we need a method that generates pairs of entities that we want to
classify as being related or not. As these candidate pairs are typically formed
within one document, this function takes a Doc as input and
outputs a List of Span tuples. For instance, a very straightforward
implementation would be to just take any two entities from the same document:
### Simple candiate generation
def get_candidates(doc: Doc) -> List[Tuple[Span, Span]]:
candidates = []
for ent1 in doc.ents:
for ent2 in doc.ents:
candidates.append((ent1, ent2))
return candidates
But we could also refine this further by excluding relations of an entity
with itself, and posing a maximum distance (in number of tokens) between two
entities. We register this function in the
@misc registry so we can refer to it from the
config, and easily swap it out for any other candidate generation function.
config.cfg (excerpt)
[model] @architectures = "rel_model.v1" [model.tok2vec] # ... [model.get_candidates] @misc = "rel_cand_generator.v1" max_length = 20
### Extended candidate generation {highlight="1,2,7,8"}
@registry.misc.register("rel_cand_generator.v1")
def create_candidate_indices(max_length: int) -> Callable[[Doc], List[Tuple[Span, Span]]]:
def get_candidates(doc: "Doc") -> List[Tuple[Span, Span]]:
candidates = []
for ent1 in doc.ents:
for ent2 in doc.ents:
if ent1 != ent2:
if max_length and abs(ent2.start - ent1.start) <= max_length:
candidates.append((ent1, ent2))
return candidates
return get_candidates
Finally, we require a method that transforms the candidate entity pairs into a
2D tensor using the specified Tok2Vec or
Transformer. The resulting Floats2 object will then be
processed by a final output_layer of the network. Putting all this together,
we can define our relation model in a config file as such:
### config.cfg
[model]
@architectures = "rel_model.v1"
# ...
[model.tok2vec]
# ...
[model.get_candidates]
@misc = "rel_cand_generator.v2"
max_length = 20
[model.create_candidate_tensor]
@misc = "rel_cand_tensor.v1"
[model.output_layer]
@architectures = "rel_output_layer.v1"
# ...
When creating this model, we store the custom functions as attributes and the sublayers as references, so we can access them easily:
tok2vec_layer = model.get_ref("tok2vec")
output_layer = model.get_ref("output_layer")
create_candidate_tensor = model.attrs["create_candidate_tensor"]
get_candidates = model.attrs["get_candidates"]
Step 2: Implementing the pipeline component
To use our new relation extraction model as part of a custom
trainable component, we
create a subclass of Pipe that holds the model.
### Pipeline component skeleton
from spacy.pipeline import Pipe
class RelationExtractor(Pipe):
def __init__(self, vocab, model, name="rel"):
"""Create a component instance."""
self.model = model
self.vocab = vocab
self.name = name
def update(self, examples, drop=0.0, set_annotations=False, sgd=None, losses=None):
"""Learn from a batch of Example objects."""
...
def predict(self, docs):
"""Apply the model to a batch of Doc objects."""
...
def set_annotations(self, docs, predictions):
"""Modify a batch of Doc objects using the predictions."""
...
def initialize(self, get_examples, nlp=None, labels=None):
"""Initialize the model before training."""
...
def add_label(self, label):
"""Add a label to the component."""
...
Before the model can be used, it needs to be
initialized. This function receives a callback
to access the full training data set, or a representative sample. This data
set can be used to deduce all relevant labels. Alternatively, a list of
labels can be provided to initialize, or you can call the
RelationExtractoradd_label directly. The number of labels defines the output
dimensionality of the network, and will be used to do
shape inference throughout the
layers of the neural network. This is triggered by calling
Model.initialize.
### The initialize method {highlight="12,18,22"}
from itertools import islice
def initialize(
self,
get_examples: Callable[[], Iterable[Example]],
*,
nlp: Language = None,
labels: Optional[List[str]] = None,
):
if labels is not None:
for label in labels:
self.add_label(label)
else:
for example in get_examples():
relations = example.reference._.rel
for indices, label_dict in relations.items():
for label in label_dict.keys():
self.add_label(label)
subbatch = list(islice(get_examples(), 10))
doc_sample = [eg.reference for eg in subbatch]
label_sample = self._examples_to_truth(subbatch)
self.model.initialize(X=doc_sample, Y=label_sample)
The initialize method is triggered whenever this component is part of an nlp
pipeline, and nlp.initialize is invoked.
Typically, this happens when the pipeline is set up before training in
spacy train. After initialization, the pipeline component
and its internal model can be trained and used to make predictions.
During training, the function update is invoked which
delegates to
Model.begin_update and a
get_loss function that calculate the loss for a
batch of examples, as well as the gradient of loss that will be used to
update the weights of the model layers. Thinc provides several
loss functions that can be used for the
implementation of the get_loss function.
### The update method {highlight="12-14"}
def update(
self,
examples: Iterable[Example],
*,
drop: float = 0.0,
set_annotations: bool = False,
sgd: Optional[Optimizer] = None,
losses: Optional[Dict[str, float]] = None,
) -> Dict[str, float]:
...
docs = [ex.predicted for ex in examples]
predictions, backprop = self.model.begin_update(docs)
loss, gradient = self.get_loss(examples, predictions)
backprop(gradient)
losses[self.name] += loss
...
return losses
When the internal model is trained, the component can be used to make novel
predictions. The predict function needs to be
implemented for each subclass of Pipe. In our case, we can simply delegate to
the internal model's predict function
that takes a batch of Doc objects and returns a Floats2d array:
### The predict method
def predict(self, docs: Iterable[Doc]) -> Floats2d:
predictions = self.model.predict(docs)
return self.model.ops.asarray(predictions)
The final method that needs to be implemented, is
set_annotations. This function takes the
predictions, and modifies the given Doc object in place to store them. For our
relation extraction component, we store the data as a dictionary in a custom
extension attribute
doc._.rel. As keys, we represent the candidate pair by the start offsets of
each entity, as this defines an entity pair uniquely within one document.
To interpret the scores predicted by the relation extraction model correctly, we
need to refer to the model's get_candidates function that defined which pairs
of entities were relevant candidates, so that the predictions can be linked to
those exact entities:
Example output
doc = nlp("Amsterdam is the capital of the Netherlands.") print("spans", [(e.start, e.text, e.label_) for e in doc.ents]) for value, rel_dict in doc._.rel.items(): print(f"{value}: {rel_dict}") # spans [(0, 'Amsterdam', 'LOC'), (6, 'Netherlands', 'LOC')] # (0, 6): {'CAPITAL_OF': 0.89, 'LOCATED_IN': 0.75, 'UNRELATED': 0.002} # (6, 0): {'CAPITAL_OF': 0.01, 'LOCATED_IN': 0.13, 'UNRELATED': 0.017}
### Registering the extension attribute
from spacy.tokens import Doc
Doc.set_extension("rel", default={})
### The set_annotations method {highlight="5-6,10"}
def set_annotations(self, docs: Iterable[Doc], predictions: Floats2d):
c = 0
get_candidates = self.model.attrs["get_candidates"]
for doc in docs:
for (e1, e2) in get_candidates(doc):
offset = (e1.start, e2.start)
if offset not in doc._.rel:
doc._.rel[offset] = {}
for j, label in enumerate(self.labels):
doc._.rel[offset][label] = predictions[c, j]
c += 1
Under the hood, when the pipe is applied to a document, it delegates to the
predict and set_annotations methods:
### The __call__ method
def __call__(self, Doc doc):
predictions = self.predict([doc])
self.set_annotations([doc], predictions)
return doc
Once our Pipe subclass is fully implemented, we can
register the
component with the @Language.factory decorator. This
assigns it a name and lets you create the component with
nlp.add_pipe and via the
config.
config.cfg (excerpt)
[components.relation_extractor] factory = "relation_extractor" [components.relation_extractor.model] @architectures = "rel_model.v1" [components.relation_extractor.model.tok2vec] # ... [components.relation_extractor.model.get_candidates] @misc = "rel_cand_generator.v1" max_length = 20
### Registering the pipeline component
from spacy.language import Language
@Language.factory("relation_extractor")
def make_relation_extractor(nlp, name, model):
return RelationExtractor(nlp.vocab, model, name)